Monday Article #59: Designing Microbial Cell Factories for the Production of Chemicals
Commercial chemicals produced from fossil resources through petrochemical refinery processes have played an integral part in human society over the past century. However, our overdependence on fossil resources and derived products have led to serious problems such as environmental pollution, extreme weather, and the depletion of fossil resources, which threatens not only humanity but also the entire planet as a whole. (Jae Sung Cho, Gi Bae Kim, Hyunmin Eun, Cheon Woo Moon, and Sang Yup Lee , 2022) Microbial cell factories (MCFs) are engineered microorganisms designed and optimized to synthesize a range of compounds to fulfil current demands of commodities in a sustainable manner. It uses a relatively lower temperature and pressure and does not use toxic solvents and catalysts for the production of chemicals, unlike conventional chemical processes. With the increasing international effort of research groups and companies around the world, a large portfolio of chemicals can now be produced using microbial cell factories. (Lee, S.Y., Kim, H.U., Chae, T.U. et.al , 2019 )
When microorganisms are first isolated from nature, they are not optimized to the desired function to readily uptake carbon sources derived from renewable biomass and produce a target chemical of interest with a high enough efficiency. Thus, metabolic engineering─the purposeful modification of cellular and metabolic networks to achieve defined objectives. (Stephanopoulos, G.; Sinskey, A. J., 1993) - is performed on the microorganism to convert it into an efficient microbial cell factory. Through metabolic engineering, a microorganism can be engineered to utilize an inexpensive renewable carbon source as a substrate to produce a chemical of interest, even those nonnative to its metabolism.
The biosynthesis pathways for the production of chemicals can be defined into three categories with reference to the microbial host selected for engineering: native-existing pathways, nonnative-existing pathways and nonnative-created pathways. Once these pathways are designed and constructed in the microbial cell factory, systems metabolic engineering strategies can be used to improve the performance of the strain to meet industrial production standards.
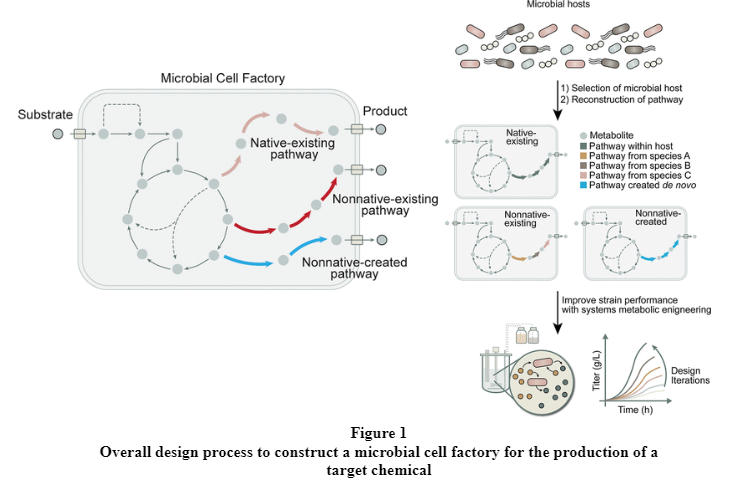
Native-existing pathways are biosynthetic pathways existing in an isolated microbial host capable of producing the target chemical endogenously without the need to introduce any foreign biosynthetic pathways. Nonnative-existing pathways are reconstructed biosynthetic pathways that utilize existing or reported pathways in nature but are nonnative to the microbial host. Nonnative-created pathways are reconstructed pathways that do not exists in nature but have purposefully designed and created using synthetic enzymes and pathways with new functions. With an increasing number of tools and strategies available to discover and design biosynthetic pathways, the production of a target chemical may not necessarily require a single category of synthetic pathway. For example, some chemicals, such as glutaric acid can be produced by both nonnative-existing pathways and nonnative-created pathways. (Jae Sung Cho, Gi Bae Kim, Hyunmin Eun, Cheon Woo Moon, and Sang Yup Lee , 2022).
Model organism Escherichia coli and Saccharomyces cerevisiae are two of the most widely used hosts for microbial cell factories because their metabolisms are best understood and molecular tools to engineer these hosts are well established. (Lee, S.Y., Kim, H.U., Chae, T.U. et.al , 2019 ) However, the production of a target chemical may not necessarily be optimal with E.coli and S. cerevisiae as hosts due to their metabolic and physiological properties. With the expanding number of genetic and computational tools available, other microorganisms with superior metabolic and cellular properties such as Corynebacterium glutamicum, Pichia pastoris, Pseudomonas putida, and Yarrowia lipolytica are increasingly being explored for the production of chemicals, with a notable increase in efforts toward the production of chemicals in the past five years. (Table 1) (Jae Sung Cho, Gi Bae Kim, Hyunmin Eun, Cheon Woo Moon, and Sang Yup Lee , 2022).

Native-Existing Pathways
There are microorganism naturally capable of producing chemicals of interest in large amounts before any metabolic engineering is performed. For example, species of C.glutamicum isolated in the 1950s were found to be capable of producing large amounts of L-glutamate and L-lysine, which are both important food and feed additives. (Eggeling, L.; Bott, M., 2005 ) These C.glutamicum isolates and their derivatives developed through classical strain improvement were directly used for the fermentative production of these amino acids. (Wendisch, V.F.; Jorge, J. M. P.; Perez-Garcia, F.; Sgobba, E., 2016) Other example include Bacillus and Lactobacillus species, which are known for the production of L-lactate, (Abedi, E.; Hashemi, S. M. B., 2020) Klebsiella pneumoniae for the production of 1,3-propanediol from glycerol, (Sun, Y.-Q.; Shen, J.-T.; Yan, L.; Zhou, J.-J.; Jiang, L.-L.; Chen, Y.; Yuan, J.-L.; Feng, E.; Xiu, Z.-L., 2018 ) Mannheimia succiniciproducens for succinic acid; (Ahn, Jung Ho , 2020 ) and actinomycetes for the production of antibiotics and polyketides. (Palazzotto, E.; Tong, Y.; Lee, S. Y.; Weber, T., 2019 )
While these native producers are advantageous for producing certain chemicals that are already inherently produced with less metabolic engineering effort, careful considerations should be paid to choosing the host for the production of a particular chemical. These include whether it harbors a strong native metabolic flux toward the synthesis of a target chemical and whether it allows chemical production with a yield close to the theoretical maximum. (Yang, D.; Park,S. Y.; Park, Y. S.; Eun, H.; Lee, S. Y. , 2020 ) Once the optimal host is chosen, metabolic pathway engineering strategies can be applied as the next step for further strain development.
Nonnative-Existing Pathways
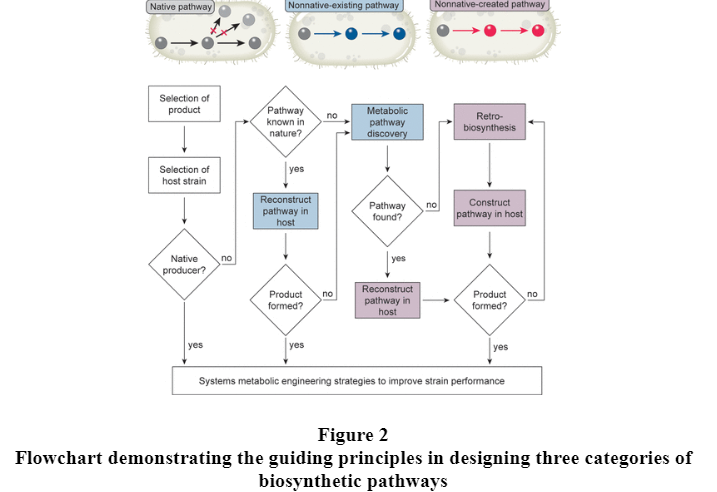
Nonnative-Existing Pathways can be further categorised into two; Reconstruction of Metabolic Pathway from Known Sources and Pathway Discovery for Unknown Metabolic Pathways.
1. Reconstruction of Metabolic Pathway from Known Sources
There are target chemicals that cannot be synthesized by a selected microbial host because the biosynthetic genes necessary for complete construction of the metabolic pathway toward a target chemical are not present in the host. This issue can be resolved because many of these target chemicals can also be synthesized by harnessing the appropriate enzymes and pathways existing in other hosts or metagenome. (Jae Sung Cho, Gi Bae Kim, Hyunmin Eun, Cheon Woo Moon, and Sang Yup Lee , 2022) Databases such as KEGG, (Kanehisa, M.; Furumichi, M.; Sato, Y.; Ishiguro-Watanabe, M.; Tanabe, M., 2021) MetaCyc (Caspi, R.; Billington, R.; Keseler, I. M.; Kothari, A.; Krummenacker, M.; Midford, P. E.; Ong, W. K.; Paley, S.; Subhraveti, P.; Karp, P. D., 2019 ) and BRENDA (Chang, A.; Jeske, L.; Ulbrich, S.; Hofmann, J.; Koblitz, J.; Schomburg, I.; Neumann-Schaal, M.; Jahn, D.; Schomburg, D., 2021 ) provide comprehensive information on metabolic pathways, enzymes and genes. ( Choi, K. R.; Jang, W. D.; Yang, D.; Cho, J. S.; Park, D.; Lee, S. Y., 2019) On the basis of this information on metabolic pathways can be reconstructed by recruiting and combining genes from other organisms or metagenome.
2. Pathways Discovery for Unknown Metabolic Pathways
There are cases where genetic or enzymatic information on certain steps in the biosynthesis pathway for target chemical production is missing. (Figure 2) Moreover, enzyme candidates for the missing biosynthetic reactions that are selected intuitively on the basis of in-depth literature searches might not function properly when introduced into the host. One way to address this issue is to discover novel enzyme candidates by profiling the genomes and transcriptomes of biosynthetic gene clusters found in nature. (Figure 3) (Jae Sung Cho, Gi Bae Kim, Hyunmin Eun, Cheon Woo Moon, and Sang Yup Lee , 2022)
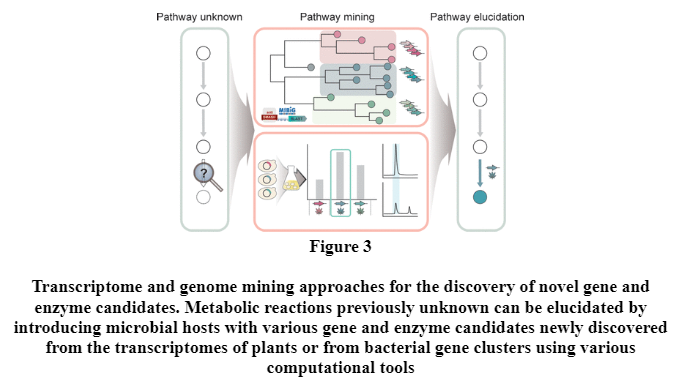
In addition, genome mining software tools such as antiSMASH, (Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S. Y.; Medema, M. H.; Weber, T., 2019 ) ClusterFinder, (Cimermancic, P.; Medema, M. H.; Claesen, J.; Kurita, K.; Wieland Brown, L. C.; Mavrommatis, K.; Pati, A.; Godfrey, P. A.; Koehrsen, M.; Clardy, J.; Birren, B. W.; Takano, E.; Sali, A.; Linington, R. G.; Fischbach, M. A, 2014 )NP.searcher ( Li, M. H.; Ung, P. M.; Zajkowski, J.; Garneau-Tsodikova, S.; Sherman, D. H. , 2009 ) and NaPDoS (Ziemert, N.; Podell, S.; Penn, K.; Badger, J. H.; Allen, E.; Jensen, P. R., 2012) enable and facilitate the discovery and prediction of cryptic biosynthetic gene clusters (BGCs) from the bacterial, fungal and plant genome sequences.
Nonnative-Created Pathways
Metabolic engineering has enabled the production of various chemicals by employing naturally existing pathways toward target chemicals. However, microbial cell factories should be capable of producing non-natural chemicals, or even natural products of which biosynthetic pathways are not characterized, to fully replace the conventional petrochemical refineries. In organic chemistry, the synthesis routes of chemicals that have no precedent cases can be designed using retrosynthesis, a discipline that works backward from a target chemical to the readily available precursors by finding appropriate chemical transformation steps. Computer-aided approaches can facilitate retrosynthesis, which can also now be used to design biosynthetic pathways, to efficiently explore the vast chemical space. Such biosynthetic pathways can be constructed de novo by identifying suitable enzymes from heterologous resources and/or harnessing the promiscuities of enzymes that are expected to catalyze the metabolite of interest. In this section, we discuss how enzyme promiscuity and retrosynthetic approaches can be applied to de novo pathway design. (Jae Sung Cho, Gi Bae Kim, Hyunmin Eun, Cheon Woo Moon, and Sang Yup Lee , 2022).
Reference(s):
Nisarg Gohil, Gargi Bhattacharjee, Vijai Singh. (2021). Microbial Cell Factories Engineering for Production of Biomolecules . Retrieved from ScienceDirect : https://www.sciencedirect.com/science/article/pii/B9780128214770000210
Lee, S.Y., Kim, H.U., Chae, T.U. et.al . (2019 ). A comprehensive metabolic map for production of bio-based chemicals. Retrieved from Nature Catalysis : https://www.nature.com/articles/s41929-018-0212-4#citeas
Jae Sung Cho, Gi Bae Kim, Hyunmin Eun, Cheon Woo Moon, and Sang Yup Lee . (2022). Designing Microbial Cell Factories for the Production of Chemicals . Retrieved from ACS Publications : https://pubs.acs.org/doi/10.1021/jacsau.2c00344#
Stephanopoulos, G.; Sinskey, A. J. (1993). Metabolic engineering-methodologies and future prospects. Retrieved from PubMed: https://pubmed.ncbi.nlm.nih.gov/7764086/
Yang, D.; Park,S. Y.; Park, Y. S.; Eun, H.; Lee, S. Y. . (2020 ). Metabolic Engineering of Escherichia coli for Natural Product Biosynthesis . Retrieved from ScienceDirect : https://www.sciencedirect.com/science/article/pii/S016777991930277X?via%3Dihub
Eggeling, L.; Bott, M. (2005 ). Handbook of Corynebacterium glutamicum. Boca Raton: CRC Press.
Wendisch, V.F.; Jorge, J. M. P.; Perez-Garcia, F.; Sgobba, E. (2016). Update on industrial production of amino acids usinf Corynebacterium glutamicum. Retrieved from PubMed : https://pubmed.ncbi.nlm.nih.gov/27116971/
Abedi, E.; Hashemi, S. M. B. (2020). Lactic acid production - producing microorganisms and substrates sources-state of art . Retrieved from ScienceDirect : https://www.sciencedirect.com/science/article/pii/S240584402031817X?via%3Dihub
Ahn, Jung Ho . (2020 ). Enhanced succinic acid production by Mannheimia employing optimal malate dehydrogenase . Retrieved from Nature Communications : https://www.nature.com/articles/s41467-020-15839-z
Sun, Y.-Q.; Shen, J.-T.; Yan, L.; Zhou, J.-J.; Jiang, L.-L.; Chen, Y.; Yuan, J.-L.; Feng, E.; Xiu, Z.-L. (2018 ). Advances in bioconcersion of glycerol to 1,3-propanedioul: Prospects and challenges . Retrieved from ScienceDirect : https://www.sciencedirect.com/science/article/pii/S1359511317318925?via%3Dihub
Palazzotto, E.; Tong, Y.; Lee, S. Y.; Weber, T. ( 2019 ). Synthetic biology and metabolic engineering of actinomycetes for natural product discovery. Retrieved from PubMed: https://pubmed.ncbi.nlm.nih.gov/30853630/
Choi, K. R.; Jang, W. D.; Yang, D.; Cho, J. S.; Park, D.; Lee, S. Y. (2019). Systems metabolic engineering strategies: Integrating systems and synthetic biology with metabolic engineering. . Retrieved from PubMed: https://pubmed.ncbi.nlm.nih.gov/30737009/
Kanehisa, M.; Furumichi, M.; Sato, Y.; Ishiguro-Watanabe, M.; Tanabe, M. (2021). Integrating viruses and cellular organisms. . Retrieved from Oxford Academic : https://academic.oup.com/nar/article/49/D1/D545/5943834?login=true
Caspi, R.; Billington, R.; Keseler, I. M.; Kothari, A.; Krummenacker, M.; Midford, P. E.; Ong, W. K.; Paley, S.; Subhraveti, P.; Karp, P. D. (2019 ). The MetaCyc database of metabolic pathways and enzymes - a 2019 update. Retrieved from Oxford Academic : https://academic.oup.com/nar/article/48/D1/D445/5581728
Chang, A.; Jeske, L.; Ulbrich, S.; Hofmann, J.; Koblitz, J.; Schomburg, I.; Neumann-Schaal, M.; Jahn, D.; Schomburg, D. (2021 ). BRENDA, the ELIXIR core data resource in 2021: new developments and updates. Nucleic Acid Research .
Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S. Y.; Medema, M. H.; Weber, T. (2019 ). antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acid Research , W81-W87.
Cimermancic, P.; Medema, M. H.; Claesen, J.; Kurita, K.; Wieland Brown, L. C.; Mavrommatis, K.; Pati, A.; Godfrey, P. A.; Koehrsen, M.; Clardy, J.; Birren, B. W.; Takano, E.; Sali, A.; Linington, R. G.; Fischbach, M. A. (2014 ). Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Retrieved from PubMed: https://pubmed.ncbi.nlm.nih.gov/25036635/
Li, M. H.; Ung, P. M.; Zajkowski, J.; Garneau-Tsodikova, S.; Sherman, D. H. . (2009 ). utomated genome mining for natural products. Retrieved from PubMed: https://pubmed.ncbi.nlm.nih.gov/19531248/
Ziemert, N.; Podell, S.; Penn, K.; Badger, J. H.; Allen, E.; Jensen, P. R. (2012). The natural product domain seeker NaPDoS: a phylogeny based bioinformatic tool to classify secondary metabolite gene diversity. Retrieved from PLOS ONE: https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0034064
This article was prepared by Emeralda Erna Nordin

Comentários